|
Research interests
Our main interest is to understand the molecular mechanisms by which oncogenes transform a normal cell into a cancerous one. We focus our research on signaling pathways that are often deregulated in metabolic diseases and cancer, such as Ras/MAPK and PI3K/mTOR.
Using high-throughput proteomic approaches to identify novel effectors of the Ras/MAPK pathway
Ras/MAPK signalling is activated by most growth factors and controls essential biological processes, including cell cycle progression, cell differentiation, survival and motility. Activating mutations in components of this pathway are frequently found in human tumours, such as in pancreatic (90%), colon (50%), thyroid (45%) and ovarian cancers (36%), as well as in melanoma (63%). We have identified a number of novel proteins regulated by the Ras/MAPK pathway, and are currently characterizing their roles in normal and cancer cells.
Assess the biological function of the RSK family of protein kinases The RSK family of protein kinases contains four human isoforms (RSK1-4). The RSK isoforms are directly activated by the Ras/MAPK pathway, but very little is know about their roles in cancer cells. Our group uses mouse genetics and molecular cell biology techniques to understand the roles of these protein kinases. We have also identified a large number of RSK substrates, which are currently being characterized as novel effectors of the Ras/MAPK pathway.
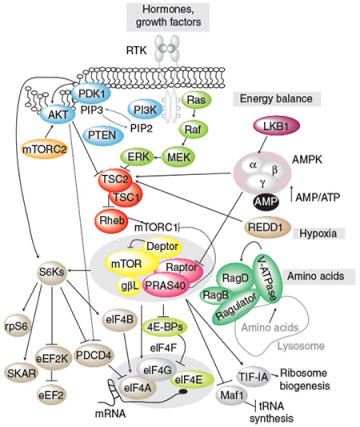
Characterize the function and regulation of the mTOR signalling pathway The mammalian target of rapamycin (mTOR) is a conserved Ser/Thr kinase that regulates cell growth as well as organ and body size in a variety of organisms. Emerging evidence indicates that deregulation of the mTOR pathway occurs in many types of cancer, underscoring the importance of understanding what lies downstream. We have identify a number of new mTOR effectors and are currently characterizing their roles in cancer cell growth and proliferation.
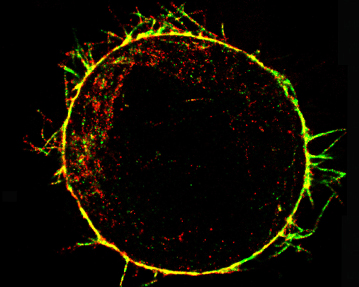
Interplay between mechanical and biological mechanisms in cell cortex assembly The cell cortex is a network of actin, myosin and associated proteins that underlies the plasma membrane and determines cell shape. As such, the cortex plays a role in normal physiology during events involving cell deformation such as mitosis, cytokinesis, and cell locomotion, and in the pathophysiology of diseases such as cancer. Despite its importance, little is known about how the cortex assembles, how it is tethered to the cell membrane, and how its mechanical properties are regulated. The aim of this project is to uncover the molecules involved in cortex assembly, understand their regulation, determine how cortex mechanical properties relate to cortical ultrastructure and proteic composition, and explore how cortical mechanics in turn influence molecular concentrations and activities. This interdisciplinary project, in collaboration with Guillaume Charras (University College London), Ewa Paluch (University of Cambridge), and Guillaume Romet-Lemonne (Institut Jacques Monod), will enable us to rationally link proteic composition, ultrastructure, mechanics and regulation of the cortex to one another.
|
Normal human mammary epithelial cells (actin filaments are stained in red, DNA in blue)
Breast cancer cells with hyperactivated MAPK signalling (as above, with the Gab2 oncogene stained in green)
Schematic of the signal transduction mechanisms stimulated by hormones and growth factors (from Roux and Topisirovic, CSHPB 2012)
Actin cytoskeleton of a rounded cell in mitosis viewed using an electron microscope (From Serres et al., Dev Cell 2020) |
.jpg)
.jpg)